1.2k by 40k (for loop takes 241 seconds, broadcasting takes only 2.5 seconds)
1.2e+03 presynaptic, 4.0e+04 postsynaptic neurons
each pre neuron has 10 connections, for 1.2e+04 total connections
---
broadcast distance calcs took
2.57 seconds
for-loop distance calcs took:
241 seconds
choosing connections took:
3.05 seconds
10k by 10k:
1.0e+04 presynaptic, 1.0e+04 postsynaptic neurons
each pre neuron has 10 connections, for 1.0e+05 total connections
---
broadcast distance calcs took
5.64 seconds
choosing connections took:
6.61 seconds

10k by 40k: only a couple of minutes!
1.0e+04 presynaptic, 4.0e+04 postsynaptic neurons
each pre neuron has 10 connections, for 1.0e+05 total connections
---
broadcast distance calcs took
63.1 seconds
choosing connections took:
28.9 seconds
The core computation of distances is done with:
#compute the difference in position by broadcasting
# https://stackoverflow.com/questions/66674537/python-numpy-get-difference-between-2-two-dimensional-array
# in theory this should be doable with something like:
# np.subtract.outer(), but I couldn't get it to works
diffs = xy_post - xy_pre[:,None] # N_pre x N_post x 2
# (x0-x1)^2 + (y0-y1)^2
dists = np.linalg.norm(diffs,axis=2)**2 # N_pre x N_post
# exp( - dist / 2sigma^2 )
gauss_fn = lambda d,sigma=SIGMA: np.exp(-d / 2*sigma**2)
probs = gauss_fn(dists)

and then connections are chosen essentially the same way you proposed:
post_idx = [[]]*N_pre
for i,row in enumerate(probs):
post_idx[i] = random.choices(range(N_post),weights=row,k=each_num_connect[i])
Notably, while this seems to be much faster than a for loop, it creates this intermediate diffs matrix which is N_pre x N_post x 2 and dists,probs which are N_pre x N_post. Your previous approach only uses 1 x N_post at a time to represent the probabilities, which should use mean less memory usage at any one point in time. 40,000 x 40,000 might be larger than what you want to store in memory at any one point in time, so you could instead use the same idea of computing all the probabilities using vector / matrix operations, but for a single presynaptic neuron. This would be like a hybrid of the for loop and broadcasting approaches (with speed and memory usage somewhere between the two).
EDIT:
here’s the entire script:
click for script
import numpy as np
import time
import matplotlib.pyplot as plt
import random
plt.rcParams['savefig.facecolor']='white'
#%%
do_plot_some = True
do_plot_all = False
do_calc_for_loop = True # set to false for large N_pre, otherwise will take 100x as much time as without
do_save_figs = False
w = 100
N_pre = 1200
N_post = 40000
SIGMA = 1.0
AVG_DIST = 5
N_conn_per_pre = 10
each_num_connect = (np.ones(N_pre)*N_conn_per_pre).astype(int)
# Create a grid of coordinates
# like: target_n_group.X, target_n_group.Y
x_pre = np.random.normal(0,1,N_pre)
y_pre = np.random.normal(0,1,N_pre)
xy_pre = np.array(list(zip(x_pre,y_pre)))
x_post = np.random.normal(AVG_DIST,1,N_post)
y_post = np.random.normal(0,1,N_post)
xy_post = np.array(list(zip(x_post,y_post)))
#%%
if do_plot_some or do_plot_all:
fig,ax = plt.subplots()
ax.axis('equal')
ax.plot(x_pre,y_pre,'k.')
ax.plot(x_post,y_post,'bx')
fig
#%%
"""
Compute gaussian distances with broadcasted vector / matrix operations
"""
a = xy_pre[0]
b = xy_post[0]
dist_fn = lambda A,B: np.linalg.norm(A-B)**2
gauss_fn = lambda d,sigma=SIGMA: np.exp(-d / 2*sigma**2)
start_broad =time.time()
#compute the difference in position by broadcasting
# https://stackoverflow.com/questions/66674537/python-numpy-get-difference-between-2-two-dimensional-array
# in theory this should be doable with something like:
# np.subtract.outer(), but I couldn't get it to works
diffs = xy_post - xy_pre[:,None] # N_pre x N_post x 2
# (x0-x1)^2 + (y0-y1)^2
dists = np.linalg.norm(diffs,axis=2)**2 # N_pre x N_post
# exp( - dist / 2sigma^2 )
probs = gauss_fn(dists)
stop_broad =time.time()
print(f'broadcast distance calcs took\n {stop_broad-start_broad:.2e} seconds')
# %%
# Check dists computation against another method
print(dists[0,0])
print(dist_fn(xy_pre[0],xy_post[0]))
#%%
"""
Compute gaussian distances with for-loop
"""
if do_calc_for_loop:
dists2 = np.zeros((N_pre,N_post))
start_for = time.time()
for i,A in enumerate(xy_pre):
for j,B in enumerate(xy_post):
dists2[i,j] = dist_fn(A,B)
probs2 = gauss_fn(dists2)
stop_for = time.time()
print(f'for-loop distance calcs took\n {stop_for-start_for:.2e} seconds')
#%%
if do_plot_all:
fig,ax=plt.subplots(2,1)
ax[0].imshow(dists)
# ax[1].imshow(dists2)
ax[1].imshow(probs,cmap='Greys')
# fig.suptitle('from')
ax[0].set_title('distances',y=0.8,color='white')
ax[1].set_title('probabilities',y=0.8,color='black')
ax[1].set_xlabel('post index')
ax[1].set_ylabel('pre index')
ax[0].set_ylabel('pre index')
if do_save_figs: plt.savefig(f'dist_prob_{N_pre}_by_{N_post}.png')
# max(dists.flatten())
fig
#%%
# fig,ax = plt.subplots()
# ax.plot(dists.flatten(),gauss_fn(dists.flatten()), 'k.')
# ax.set_xlabel('squared distance')
# ax.set_ylabel('probability: gaussian(sqr dist)')
# fig
#%%
post_idx = [[]]*N_pre
start_choose=time.time()
for i,row in enumerate(probs):
post_idx[i] = random.choices(range(N_post),weights=row,k=each_num_connect[i])
stop_choose=time.time()
print()
print(f'choosing connections took: \n {stop_choose-start_choose:.2e} seconds')
#%%
if do_plot_some or do_plot_all:
fig,ax=plt.subplots()
ax.axis('equal')
# Plot pre-synaptic neurons
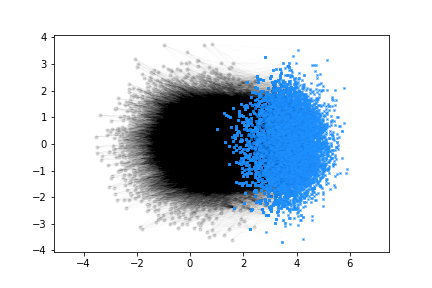
ax.plot(x_pre,y_pre,'.',color='lightgrey')
for i in range(N_pre):
for j in post_idx[i]:
ax.plot([x_pre[i],x_post[j]],[y_pre[i],y_post[j]],'-',color='black',alpha=0.1,linewidth=.1)
#NOTE: if N_post is big, only plot markers for connection targets
ax.plot(x_post[j],y_post[j],'x',color='dodgerblue',markersize=2)
if do_plot_all:
# Plot post-synaptic neurons
ax.plot(x_post,y_post,'x',color='dodgerblue',markersize=2)
if do_save_figs: plt.savefig(f'connections_{N_pre}_by_{N_post}.png')
fig
#%%
# Print summary report
print(f'{N_pre:.1e} presynaptic, {N_post:.1e} postsynaptic neurons')
print(f' each pre neuron has {N_conn_per_pre} connections, for {N_conn_per_pre*N_pre:.1e} total connections')
print('---')
print(f'broadcast distance calcs took\n {stop_broad-start_broad:.2e} seconds\n')
print(f'for-loop distance calcs took\n {stop_for-start_for:.2e} seconds\n')
print(f'choosing connections took: \n {stop_choose-start_choose:.2e} seconds\n')
I use Hydrogen in a similar way to a jupyter notebook, where #%% denotes a new cell, but this is also runnable as a regular python script.
-Adam



